Story highlights
A new technique allows ancient DNA to be studied with specific results
"Pestilence" cemetery contained salmonella bacteria, which likely caused typhoid fever
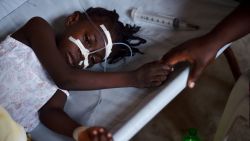
In the 16th century, an epidemic known as “cocoliztli” that caused bleeding and vomiting swept through large areas of Guatemala, Mexico and even reached Peru. It wiped out 80% of the population, killing millions of people.
Ancient DNA and a new technique have been used to determine the likely cause of this mysterious epidemic that contributed to a “cataclysmic” population decline.
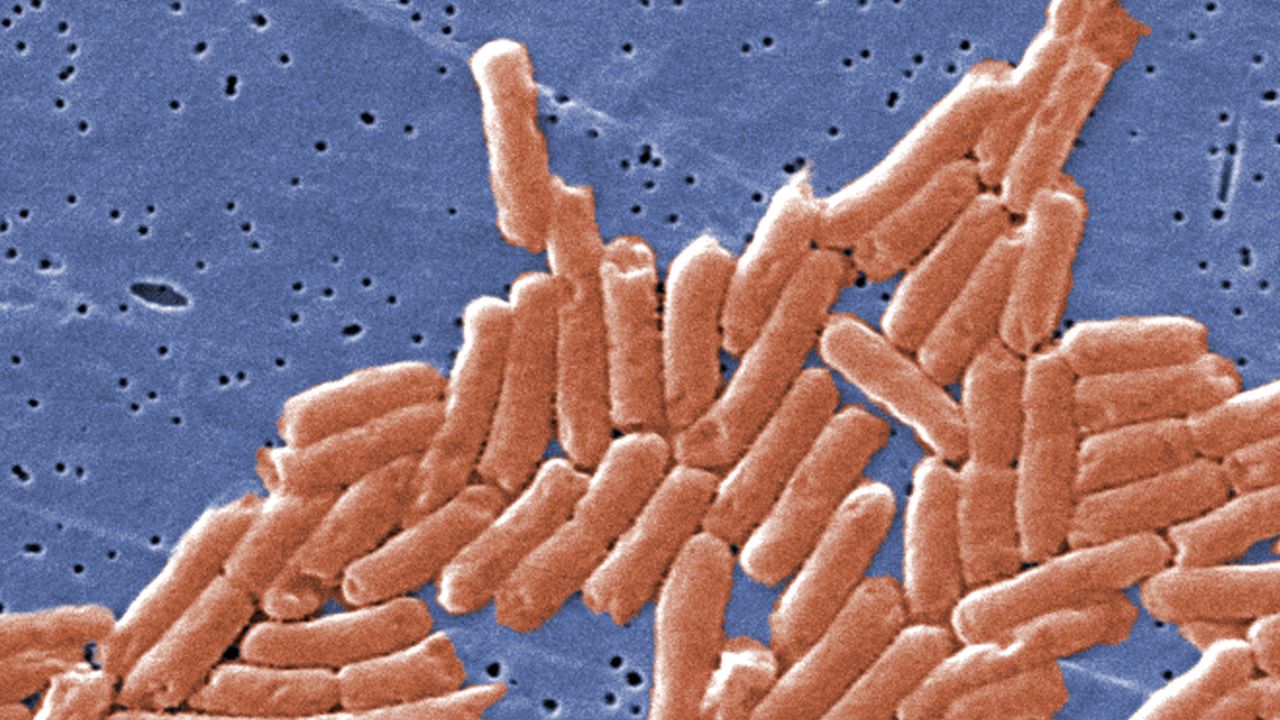
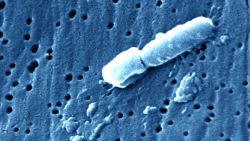
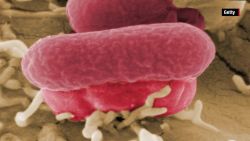
Salmonella genomes, which cause typhoid fever, were recovered from DNA within the teeth of 10 skeletons buried in an undisturbed “cocoliztli” or “pestilence” cemetery in Oaxaca, Mexico. This would be the first known occurrence of salmonella in the Americas, according to a new study published in the journal Nature on Monday. Typhoid fever has long been suspected due to the recorded symptoms, but this is the first identification of bacteria at the site.
The researchers also believe that the arrival of Europeans to what was then known as Mesoamerica caused the devastating epidemic. Europeans were susceptible to enteric fever, also known as typhoid fever, and it is very likely that they were carriers for the disease when they arrived to conquer Mesoamerica.
“The cocoliztli is a mysterious historical epidemic, and over the years many have speculated on its cause,” said Kirsten Bos in an email, study author and group leader of molecular palaeopathology at the Max Planck Institute for the Science of Human History in Germany. “This is the first time that ancient DNA has been successful in identifying a candidate pathogen for it.”
The Teposcolula-Yucundaa’s Grand Plaza cemetery is the only one known to be linked to this specific outbreak. The epidemic was so devastating that the city was relocated to a nearby valley, which allowed the cemetery to remain untouched for centuries. This, along with the thick, protective floor of the Grand Plaza, created the perfect conditions for testing and research.
While the culprit diseases behind later epidemics, like smallpox, measles, mumps and influenza, have been well documented, earlier epidemics in the “New World” aren’t as well-characterized, creating debate among researchers.
Infectious disease pathogens don’t leave telltale marks on skeletons, either, according to the researchers. This is largely due to the fact that they are fast-acting and take their toll very quickly before the skeleton can be deformed in any way.
So when researchers look at skeletons like those in the pestilence cemetery, they have to search for possible causes based on what they know from historical accounts. But diseases and symptoms can change over the years, or the symptoms can be so broad and similar that it could be one of many causes.
But a new screening technique called the Metagenome analyzer Alignment Tool, or MALT, allowed the researchers to search for all bacterial DNA present, rather than testing for each specific possibility – which can be tedious and disappointing. It’s the classic “needle in a haystack” scenario.
Other factors can also play a role. When archaeological tissues sit in the ground for centuries, DNA from environmental sources can leach in, Bos said.
“One limitation that we, and everyone else, face is that we can only look for pathogenic organisms that we already know exist and that have been genetically characterized today,” said Åshild Vågene in an email, study author and doctoral student in the department of archaeogenetics at Max Planck Institute for the Science of Human History. “If the individuals that we studied from Oaxaca, Mexico, were infected by something that does not exist today or that has yet to be characterized, then we would not be able to detect it with out current method.”
MALT revealed Salmonella enterica Paratyphi C, the bacterial cause of enteric/typhoid fever, which has been the suspected cause of the epidemic for years. Identifying the bacteria supports the typhoid hypothesis. Symptoms of typhoid fever include high fevers, red spots dehydration, bleeding, vomiting and gastro-intestinal issues.
“After we identified traces of Salmonella enterica DNA using our new computational technique, we conducted further experiments and computational analyses that allowed us to study the whole genomes of the Salmonella enterica bacteria identified in the teeth of individuals included in our study,” Vågene said.

But the case may not be closed.
“We cannot say that it definitively caused the epidemic,” Bos said. “It was the only pathogen that surfaced from our extensive analysis, and an enteric fever is consistent with the recorded symptoms of the epidemic. But it may not have been the only disease circulating in the population at this time. Others could have been present that were not detectable by us through the techniques we used.”
Follow CNN Health on Facebook and Twitter
The MALT technique is opening up new research possibilities for diagnosing diseases of the past, and solving centuries-old medical mysteries.
“The screening technique used here will be transformative for future work on archaeological disease – it’s no longer necessary to have a candidate pathogen in mind for molecular detection,” Bos said. “The flexibility offered by our approach is what’s needed to tackle many questions related to disease history and ecology, where you often don’t know what disease you’re looking for until you’ve found it.
“We intend to apply similar techniques to search for diseases in other archaeological samples from different time periods and locations. This technique opens so many doors for us to learn about disease in the past.”