CNN Labs explores the latest innovations around the world to showcase cutting-edge designs and groundbreaking research in science and technology.
Story highlights
Genetic admixure: When people from different groups interbreed
There have been many instances of this happening
Researchers used DNA to track historical mixing
Most people are carrying around historical records in their DNA, with clues to how people from genetically distinct groups intermingled over the last 4,000 years. That’s the conclusion of a new study in the journal Science.
Researchers set out to pinpoint which groups intermixed, and when, over thousands of years, using modern DNA. They thought this information could complement other historical records.
“The results suggest DNA is indeed a powerful means of reconstructing history, which then might be used to identify the genetic legacies of historically well-attested events … and highlight surprising events with a less clear historical context,” authors Garrett Hellenthal and Simon Myers wrote in an e-mail.
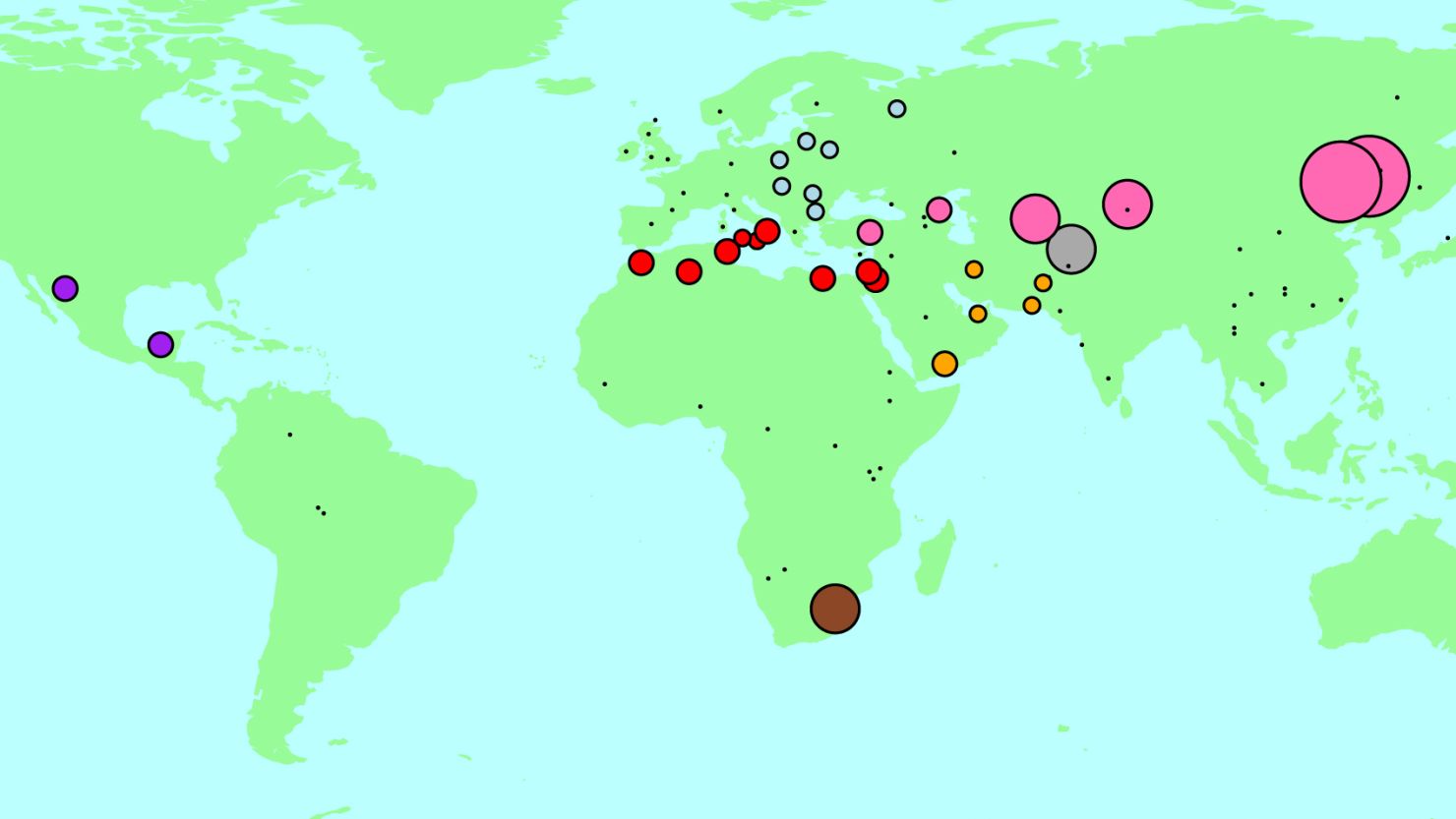
The scientists have created an interactive map showcasing their findings. You can explore the map in detail on the researchers’ website, admixturemap.paintmychromosomes.com.
The technical term for people from genetically different groups having babies together is “genetic admixture.” This has happened throughout history when people migrated into a new geographic region where others were already living.
Tell us your story!
Tell us your story!
We love to hear from our audience. Follow @CNNHealth on Twitter and Facebook for the latest health news and let us know what we’re missing.
Researchers say there have been many such admixture events among people from populations that were separated by thousands of miles. Sometimes populations adjacent to each other have distinct ancestry and history.
Did you know that your DNA contains genetic segments you’ve inherited from groups that intermingled in the past? Researchers can look at these signatures for clues about major historical mixings of populations.
DNA from a person today can also give scientists an idea of how long ago two groups genetically mixed.
The more time that has passed since the admixture event, the shorter the inherited DNA segments from the original groups are likely to be. That’s because there would have been more generations in between, and there would have been reshuffling of genetic material in each generation.
There are limitations to the method, however. When genetic mixing involves more than three groups, the researchers can’t identify those specific events. The researchers were also limited in how well they could estimate when the same, or very similar, groups interbred on multiple occasions in one descendant group’s history.
Oldest human DNA found in Spain
The researchers looked at a genetic data set of 1,490 people who come from 95 genetically distinct groups worldwide. Their software, Globetrotter, was able to describe genetic mixing events dating back about 4,500 years.
The expansion of the Mongol empire, which took place from 1206 to 1368, appears to have been highly influential on many populations in Europe and Asia. Previous studies have also supported that idea. Genghis Khan and his armies began this monumental historical event that influenced seven populations included in this study.
The Arab expansion and slave trade, which began around the seventh century, also had a large genetic impact on many groups, including people from around the Mediterranean, the Arabian Sea and the Persian Gulf. The study’s findings are consistent with that historical timeframe, but suggest that sub-Saharan Africans and Moroccans mixed earlier.
A group called the Kalash, from Pakistan, appears to come from an admixture event that happened sometime before 206 B.C., when people from a more European group hooked up with a more Central/South Asian group.
Researchers say the Kalash are geographically isolated, originating in the Hindu Kush mountains, which is why they don’t show genetic signatures of a more recent admixture event. Local tradition says the Kalash are descended from the army of Alexander the Great; the new study does not rule that out, since the time period of the mixing event could have been about right.
“We have tried to let the genetic information ‘speak’ in this study, but future interpretation of signals – especially in follow-up studies of specific regions – would benefit from expertise from … historians, linguists and others,” Hellenthal and Myers said.
Some groups showed genetic signals that the researchers were unable to precisely interpret. For instance, genetic material from French people suggests an intermixing between northern and southern Europeans and northern Africans, between 785 and 1,385 years ago, but the event itself is still unclear.
The researchers were not able to pick up signals of some well-established historical intermixings, such as migrations into the United Kingdom from northern European groups, because of the small sample sizes.
It is possible that a mutation could arise in two populations independently, but that is thought to be rare.
“If two individuals share a mutation, it is statistically much more likely that the mutation occurred in a common ancestor of both individuals, rather than arising independently in the ancestors of each of them,” Hellenthal and Myers said.
The researchers are involved in a project called People of the British Isles, which is looking specifically at the genetic origins of people from the United Kingdom. Other projects are looking in more depth at areas such as the Caribbean, Ethiopia and the Americas.
The researchers say in the study that as whole-genome sequencing improves and analyses are able to incorporate more people as well as ancient DNA, we may reach a “better understanding of ancient events where little or no historical record exists, to identify many additional events … and to provide more precise event characterization than is currently possible.”
There may also be implications for public health, as researchers get better at identifying disease risks that are higher in specific populations.